Diseases such as polio, the common cold, and meningitis are all caused by closely related viruses, and the way these viruses multiply in the body is poorly understood. Deepak Koirala, assistant professor of chemistry and biochemistry at UMBC, has already begun to unravel the mystery. Now, with a $786,000 NSF CAREER Award, his research group will be able to answer even more questions. In particular, they will investigate the RNA structures within the genetic material in these viruses and how those structures enable the viruses to multiply inside cells. The answers could eventually lead to drugs that attack specific mechanisms critical for viral replication, stopping these diseases in their tracks.
Koirala’s lab works on enteroviruses, a group of viruses that have a genome made of a single strand of RNA, rather than double-stranded DNA (like in humans). “RNA is a really versatile and dynamic molecule that functions in pretty much every aspect of cellular processes,” Koirala says.
In viruses with RNA genomes, the genome must control both the process that copies (replicates) the genome and the process that converts the genetic code into proteins. Both processes involve coordination of numerous viral and host cell proteins. The RNA must also somehow continually “decide” between the two processes. In DNA genomes, the DNA is only responsible for replication.
With the new grant, Koirala’s group seeks to better understand how enteroviruses make the decision between copying their genome and building proteins. But before they can do that, they need to nail down the three-dimensional RNA structures within the enterovirus genome that are involved in those processes.
Defining the target
Based on the way enterovirus genomes behave in biochemical studies, previous research has predicted that the beginning of an enterovirus’s genomic RNA strand folds up on itself, forming a shape resembling a cloverleaf. That structure builds a platform to assemble the viral and host proteins required for replication. This idea is widely accepted, but the precise three-dimensional structure of this region, the so-called “cloverleaf RNA domain,” and how it regulates viral replication is unknown. Figuring that out is the Koirala lab’s main task.
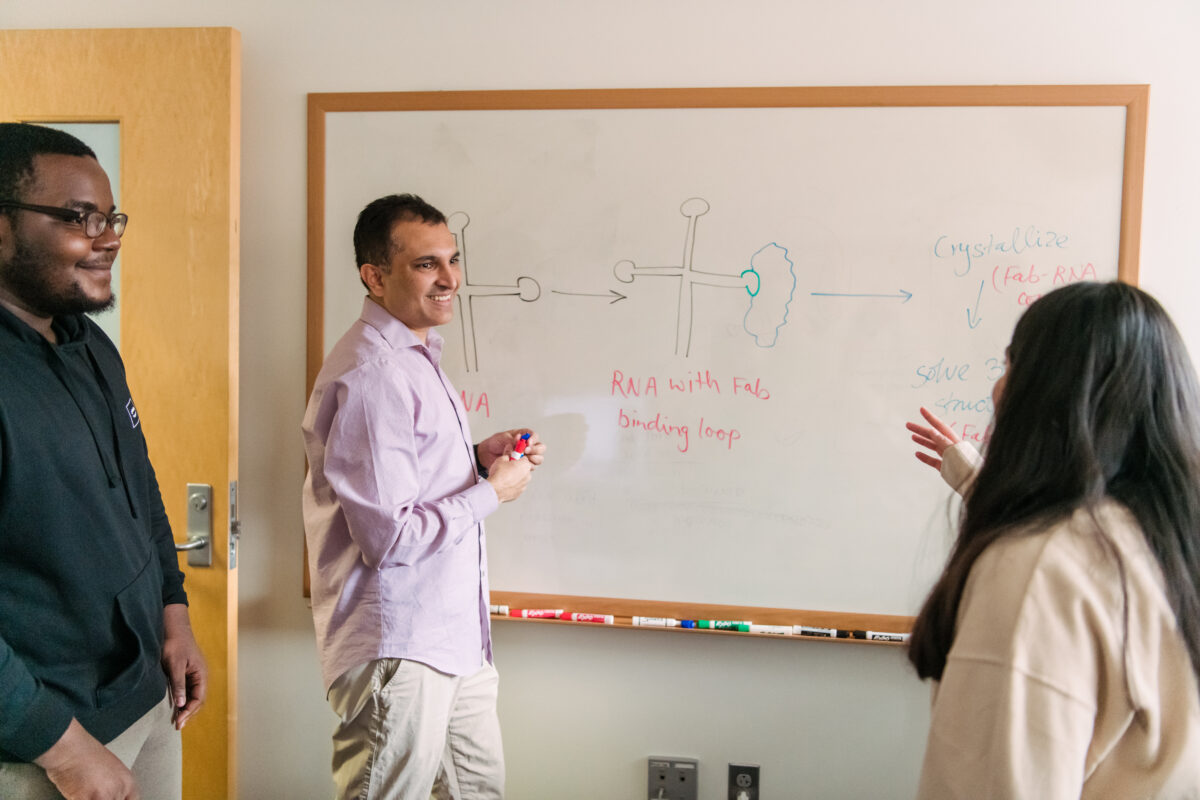
They are well on their way. Koirala’s group recently determined the cloverleaf structure from a coxsackievirus, which causes hand-foot-and-mouth disease, and is an important model system for studying many other human viruses. It will be published in a forthcoming paper in Nature Communications. “I think the field will be really excited to see this,” Koirala says. “It would be the first three-dimensional structure of the full-length cloverleaf domain for the entire enterovirus genus.”
Because the cloverleaf domain is so important for viral replication, the expectation is that its structure will be similar, if not identical, across all enteroviruses. With the new grant, Koirala hopes to determine the 3D structures of this region in several more enteroviruses. The structures the sequences form that are the same or similar across species are most likely to play similar key roles in the viral life cycle.
“That will create the opportunity to get a generic target that might be able to treat more than one of these viruses,” Koirala says. “If you really hit a structure in coxsackievirus, for example, that’s shared across many other enteroviruses, then that could be equally useful for, say, rhinovirus. In the long term, that could be really powerful.”
Crystals and X-rays
Determining the 3D structure of RNA is notoriously difficult. Koirala’s group uses a technique called X-ray crystallography, where one must first turn the RNA into a crystal through a laborious process. Then a machine directs X-rays through the crystal and then a detector records the reflections that come out. By examining those reflections, called diffraction patterns, the researchers can deduce the molecule’s shape in the crystal. Then, they map the known sequence of RNA bases onto the shape for a final 3D structure.
To make this a little easier, Koirala’s group uses a cutting-edge technique that results in successful crystallization more often than traditional methods. Koirala came to UMBC in 2020 after completing a postdoctoral fellowship in the research group at the University of Chicago that pioneered the technique.
The technique involves attaching a fragment of a synthetic antibody to the RNA, which serves as a “chaperone” to help the RNA crystallize. RNA is coated with negative charges, which repel each other and make it harder to pack the molecules tightly together—a necessary part of crystal formation. When the RNA binds to the protein, those negative charges are neutralized. And, because the protein’s structure is known, that makes it easier to detect the unknown RNA structure in the crystal.

“Just the beginning”
But even after all that, “The structure is just the beginning,” Koirala says.
RNA does not normally exist as crystals. Therefore, for one, it is important to know if the 3D structure of the protein-bound, crystallized RNA accurately represents what the RNA looks like in a biological context. But with the crystal structure in hand, “Now we have more idea about what to do next,” Koirala says, “to show what the important features of that particular structure are that dictate or define the function.” Follow-up biochemical experiments with deliberately modified versions of the RNA can help tease out which parts of the structure are critical for different functions.
And finally, Koirala says, “Now, with a well-characterized RNA structure, one has an opportunity to design a drug molecule, for example, that precisely targets that RNA structure and stops the genome replication.”
A strong team
With the new funding, Koirala will be able to grow his already sizable team. That way he can accomplish more in the lab—and also expose more students to research. Koirala is happy to bring on UMBC freshmen and sophomores as well as students with more research experience. Even local high school students have gotten involved.
“If you expose students to research early on, that gives them the opportunity to decide which career will work for them,” Koirala says. And by getting students involved right away, they are apt to stay in the lab for a few years—enough time to significantly grow their skills and even become authors on a scientific paper, he explains.

In addition to conducting their own research, the students “also get great experiences with mentoring,” Koirala says. With a group of 11 students (“Or 12, including me,” Koirala adds), a mentorship structure forms naturally among the lab members, with more experienced team members guiding and supporting newer team members. “And wherever they go, academia or industry, they will be the future scientists—they will mentor the younger ones.”
Koirala’s method seems to be working. Two high school students in the lab are headed to college next year, one to M.I.T. and one to Bowdoin College in Maine. Tasnia Sadat ’23, biochemistry and molecular biology, is headed to medical school at Georgetown. Jeff Vogt ’23, biochemistry and molecular biology, is on his way to Johns Hopkins for a Ph.D., and Senali Dansou ’23, biochemistry and molecular biology, will matriculate at University of Minnesota for an M.D.-Ph.D.
With such an engaged group of researchers, thoughtful research questions, and effective techniques, Koirala’s team is well prepared to reach its goals and further the understanding of enteroviruses, leading the way for life-changing treatments.
Tags: ChemBiochem, CNMS, majoraward, NSF CAREER award, Research

