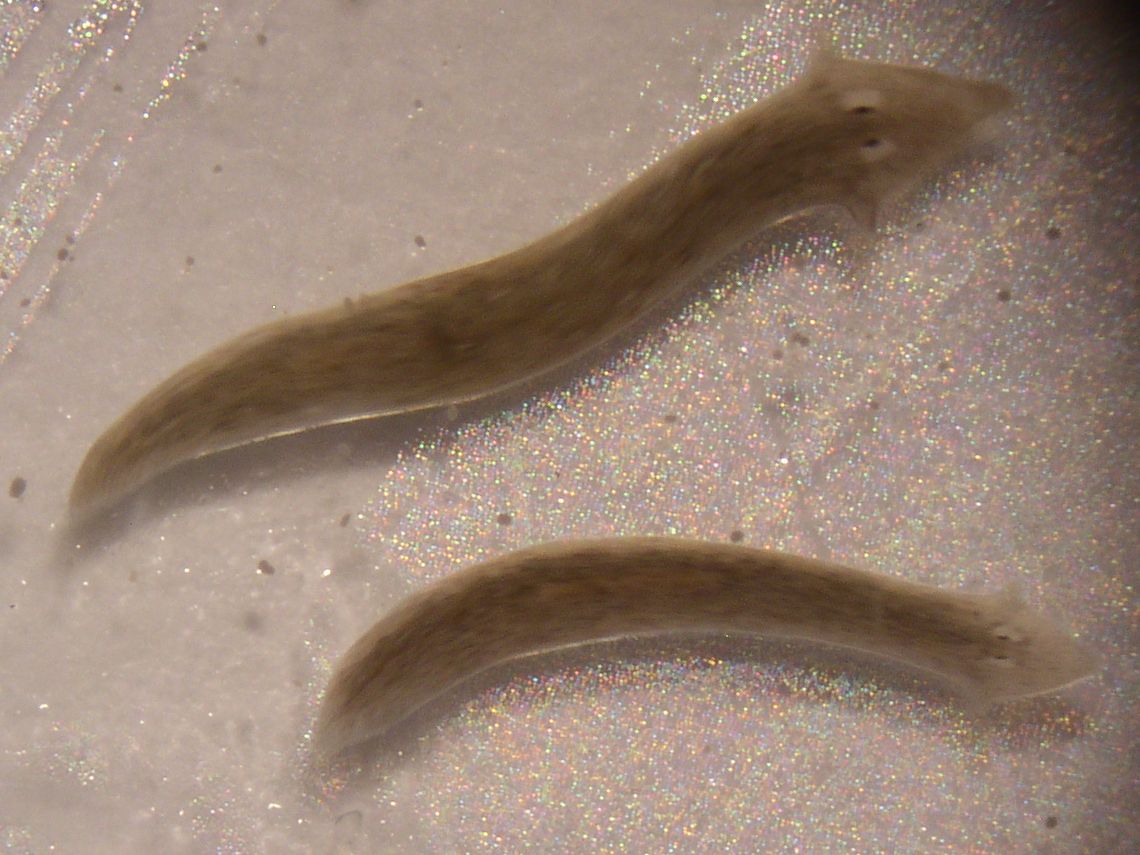
Salamanders regenerate their tails. Sea stars regenerate their arms. Most species of planaria, a type of flatworm, can regenerate everything from their heads (complete with brain) to their digestive organs. But if you lose part of a finger in a shop class accident, or while chopping vegetables for dinner, you’re out of luck—for now.
“Why can the worm do it, and we cannot?” asks Daniel Lobo, assistant professor of biological sciences. That’s not really the question, though, he explains.
“We were able to generate ourselves when we were embryos. So we have all the information of how to generate a new hand, for example,” Lobo points out. “The genes are there. You have the same information in your cells.” So why can’t humans generate body parts after that early stage of development?
“Actually, we could,” Lobo argues, if we could somehow reactivate the same genes that enabled us to develop in the womb. So, “Can we reactivate them?” he asks. That’s the real question, which he is working to answer with a five-year, $1.9 million grant from the National Institutes of Health.

Restoring independence
Lobo is tackling this question through a unique combination of techniques: wet lab experiments with planaria, and machine learning approaches that use computers to help deduce genetic regulatory networks. Previous work successfully restored the regeneration capacity of a species of planaria that had lost that ability. While still a long way from growing a human finger back, it’s a sign that the promise of reclaiming regeneration is not so far-fetched.
This line of research could eventually make it possible for people with limb loss, such as injured veterans, to regrow lost body parts. By increasing understanding of genetic regulation, Lobo’s work might also enhance knowledge of development and developmental diseases, and how cancerous tumors work around regulatory networks to grow unchecked.
The right worm for the job
Lobo uses an approach known as systems biology to tackle these big ideas. “We mix the fields of math, computer science, and biology,” he says. “We use computational techniques to extract knowledge from biological data sets.” The result is mathematical models that can explain observations the team makes in the lab. The models can also make predictions, which researchers can test in the lab.
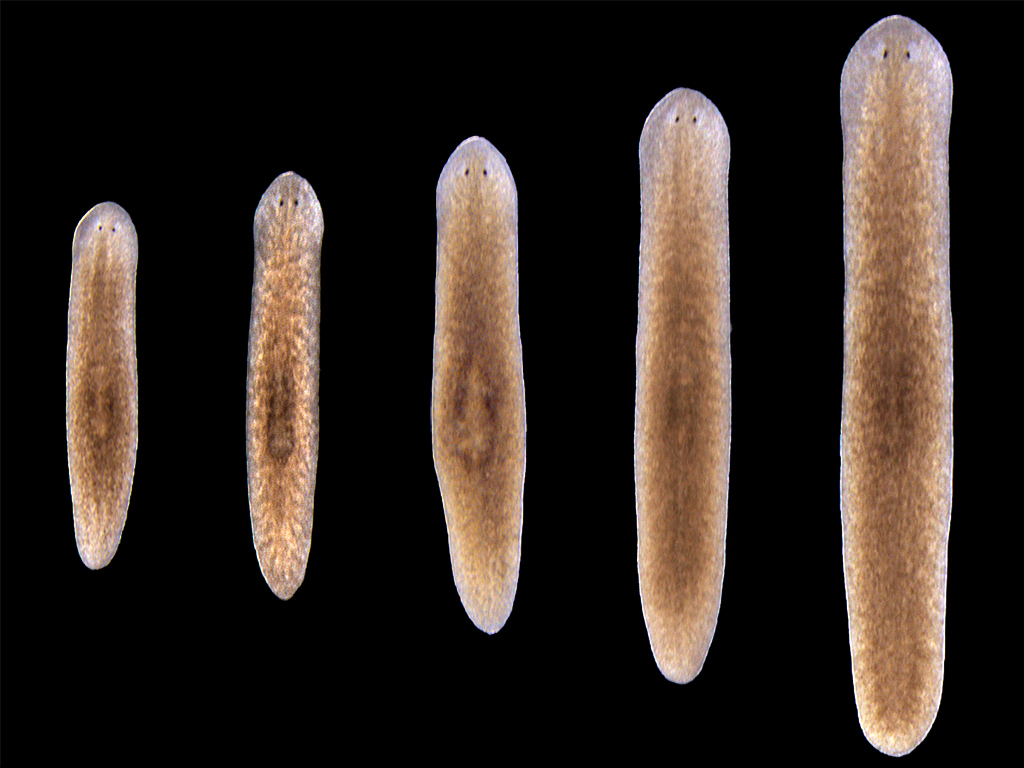
Planaria are the ideal model organism for lab work, because of their astonishing ability to regenerate. Even a worm in eight pieces will grow back into eight complete worms with proper proportions. Like mammals, the worms also grow when they have enough to eat. However, when hungry, rather than simply getting thinner, their whole bodies shrink to maintain proper proportions. So, beyond regeneration, “The general idea is to understand how gene regulation works to specify shapes and forms in biology,” Lobo says.
Teamwork and flexibility
“This program is too hard to do with just wet lab or just computational approaches,” Lobo says. “You need both.”
Because the work requires such a range of techniques, it also requires team members with a range of skills. Lobo’s lab includes undergraduate and graduate students in math, computer science, bioinformatics, and biology. The new grant will also allow him to bring on two new postdoctoral fellows, one on the wet lab side and one computational.
“We will be able to create that synergy and get people trained in both fields in the same lab,” says Lobo. He describes the interactions between lab members from different fields as essential to the success of the research.
Rather than fund a specific project, Lobo’s new Outstanding Investigator Grant will fund the lab as a whole. “It gives you a lot of freedom to adapt to whatever discoveries you make,” he says. “You have the flexibility to pursue the details that you need to.”
Teaching computers so they can teach us
The computers the team uses are powerful, but for now, they still benefit from some human guidance. To give the computers a head start on figuring out the genetic regulatory networks, the team inputs certain rules before they add loads of data from their own experiments and other labs’ work. That also ensures the computers don’t come up with a solution that is biologically impossible.
“We know that genes generally interact in certain fashions, and those interactions can be represented in different ways mathematically,” Lobo explains. “So we can tell the computer what kinds of interactions a gene can have. And then it is free to put those interactions together in ways that make sense.”
Lobo compares it to working with Lego blocks. “How many structures can you make with Legos? Unlimited, right?” he asks. “So the computer also has an unlimited space to search, but only with things that can be put together. You cannot make a perfectly round Lego ball, for example, if you only have square blocks.”

Speeding up the science
Combining wet lab and computational approaches will drastically increase the pace of discovery. A high performance cluster of computers can come up with a probable solution by testing more than a billion possible models of a regulatory network in a few days—a task that would take infinitely long for a team of humans.
Using computers to come up with plausible models, testing the models’ predictions in the lab, and then feeding the new data back into the computers to refine the model will bring researchers ever closer to understanding how different biological systems work. Research teams can apply the same investigative process to any number of biological questions, from regeneration to metastasis.
Some biologists may shy away from programming, but as Lobo says, “Biology is more and more computational. We are reaching a point that without a computer to process the data you cannot do almost any experimental work in biology.”
He argues that interdisciplinary teams like his are the future—that diverse groups of researchers will increasingly combine multiple approaches to answer the big questions, to speed up scientific progress in ways that will have real, positive impact.
Banner image: Daniel Lobo in front of the Biological Sciences Building mural. Photo by Marlayna Demond ’11 for UMBC.