A new paper in Nature Communications illuminates how a previously poorly understood enzyme works in the cell. Many diseases are tied to chronic cellular stress, and UMBC’s Aaron T. Smith and colleagues discovered that this enzyme plays an important role in the cellular stress response. Better understanding how this enzyme functions and is controlled could lead to the discovery of new therapeutic targets for these diseases.
The enzyme is named ATE1, and it belongs to a family of enzymes called arginyl-tRNA transferases. These enzymes add arginine (an amino acid) to proteins, which often flags the proteins for destruction in the cell. Destroying proteins that are misfolded, often as a result of cellular stress, is important to prevent those proteins from wreaking havoc with cellular function. An accumulation of malfunctioning proteins can cause serious problems in the body, leading to diseases like Alzheimer’s or cancer, so being able to get rid of these proteins efficiently is key to long-term health.
Tantalizing implications
The new paper demonstrates that ATE1 binds to clusters of iron and sulfur ions, and that the enzyme’s activity increases two- to three-fold when it is bound to one of these iron-sulfur clusters. What’s more, when the researchers blocked cells’ ability to produce the clusters, ATE1 activity decreased dramatically. They also found that ATE1 is highly sensitive to oxygen, which they believe relates to its role in moderating the cell’s stress response through a process known as oxidative stress.
![At left, two different molecular structures, represented by red and yellow balls connected by black lines, labeled "[4Fe-4S]" and "[2Fe-2S]". A silver arrow points from the structures to an orange blob labeled ATE1. Below the orange blob, there is a blue blob to the left labeled "tRNA-Arg." It has a small green dotted-line circle with an R inside attached to it. Below right of the orange blob, there is another blue blob, labeled "substrate." It also has a green circle with an R inside attached to it. An arrow passing from the blue blob on the left to the blue blob on the right is labeled "Arginylation."](https://umbc.edu/wp-content/uploads/2023/03/FeSabstract-v24-1200x600.png)
“We were very excited about that, because it has lots of very tantalizing downstream implications,” particularly related to the enzyme’s role in disease, says Smith, associate professor of chemistry and biochemistry.
Smith’s lab works initially with the yeast protein but also showed that the mouse version of ATE1 behaves similarly. That’s important, Smith explains. “Since the yeast protein and the mouse protein behave the same way,” he says, “there’s reason to believe, that because the human protein is quite similar to the mouse protein, it likely behaves the same way as well.”
A new approach
Before they made their breakthrough discovery, Smith and then-graduate student Verna Van, Ph.D. ’22, biochemistry and molecular biology, had been attempting for quite some time to induce ATE1 to bind with heme, a compound that contains iron and is necessary to bind oxygen in blood, to confirm another group’s results. It wasn’t working, and they were getting frustrated, Smith admits. But one day, as Smith was preparing a lecture on proteins that bind with clusters of metal and sulfur atoms, he realized the proteins he was about to cover with his students looked similar to ATE1.

After that realization, Smith and Van took a new approach. In the lab, they added the raw materials for creating iron-sulfur clusters to a solution with ATE1, and the results showed that ATE1 did indeed bind the clusters. “This looks promising,” Smith remembers thinking. “We were super excited about it.”
The fact that the enzyme binds the clusters at all was interesting and new, “but then we also asked if that’s affecting the enzyme’s ability to do what it does,” Smith says. The answer, after more than a year of additional experiments, was a resounding yes. In the process, Smith’s group also determined the structure of ATE1 in yeast (without the cluster bound to it), which they published in the Journal of Molecular Biology in November 2022.
Subtle but significant
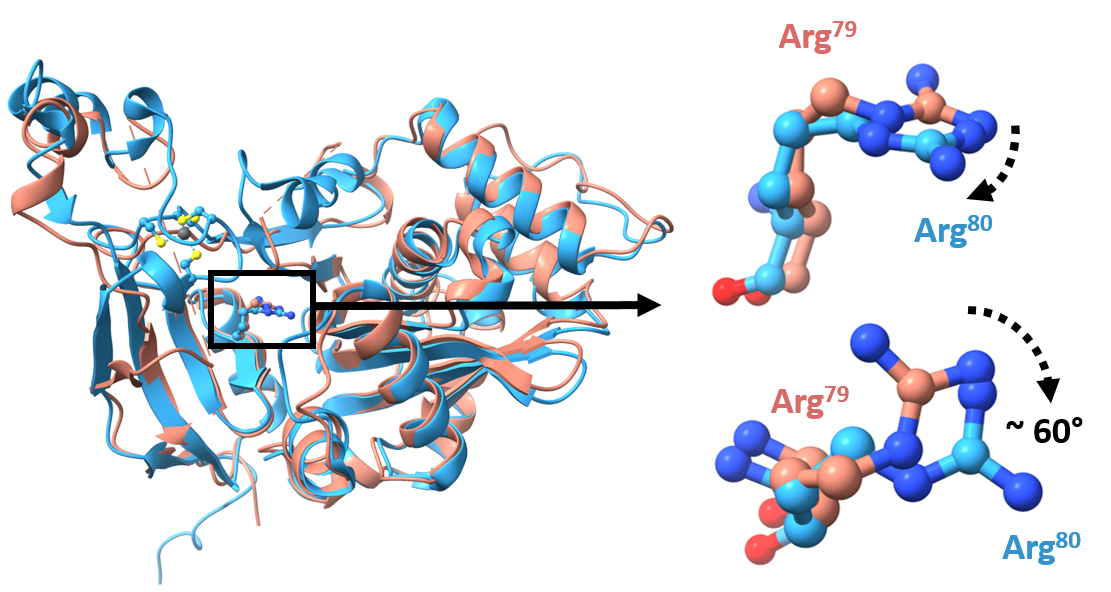
Around the same time, another group also published a slightly different ATE1 structure. The other group’s structure had a zinc ion (another metal) bound in place of the iron-sulfur cluster. With the zinc in place, one key amino acid is rotated about 60 degrees. It might seem inconsequential, but Smith believes that rotation, which he presumes is similar with the cluster, is the key to the cluster’s role in ATE1’s function.
The rotated amino acid is directly adjacent to where a protein would interact with ATE1 to be modified, ultimately flagging it for degradation. Changing the angle of that amino acid changes the shape of the location the protein would bind “very subtly,” but changes its activity “more than subtly,” Smith says.
Looking ahead and looking back
Smith would also like to explore how other metals, beyond zinc and the iron-sulfur cluster, may affect the enzyme’s activity. Additionally, his lab is working to determine the structure of ATE1 in an organism other than yeast and to confirm the ATE1 structure with an iron-sulfur cluster bound.
All these steps will build up a clearer picture of how ATE1 functions and is regulated in the cell. Smith also says he believes proteins that so far have not been shown to bind iron-sulfur clusters may indeed rely on them.
This new paper actually harks back to Smith’s first days at UMBC. He has always been interested in protein modifications, and adding arginine is a more unusual one. “It’s always something that I had filed back in my mind, and thought, ‘Oh, it would be really interesting to get a better understanding of how that works,’” he says.
Several years later, his group is now on the leading edge of discovering how arginine modifications influence cellular function and disease.
Tags: ChemBiochem, CNMS, Research