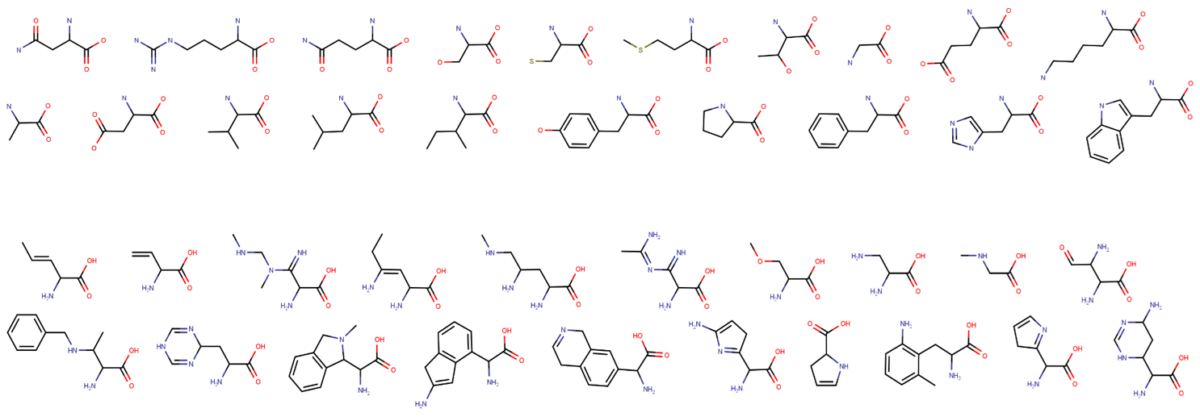
Every living thing on Earth, from bacteria to humans, uses the same set of 20 amino acids to build all of its proteins, called our “amino acid alphabet.” But why that particular set of 20? If other amino acids are possible, what makes a good alphabet? And how might we recognize life beyond Earth that’s using an alphabet other than our own?
These are all questions that Stephen Freeland, professor of biological sciences, has been asking for over a decade in his theoretical research at UMBC. Now he and colleagues at the Tokyo Institute of Technology, Charles University in the Czech Republic, and Johns Hopkins University will take this work into the laboratory for the first time.

The team seeks to marry Freeland’s theoretical work with his colleagues’ unique laboratory expertise to link together known alternative amino acids to form proteins. The Human Frontier Science Program (HFSP) is supporting the work with a new three-year, $1.8 million grant. The HFSP is a major international funding agency for basic life science research that specializes in supporting collaborations across continents.
The overarching question the project seeks to answer, Freeland says, is “What can we learn about how life could be different at the most fundamental level of biochemistry?” It’s a daunting question, with significant implications from biomedical applications to increasing our chances of detecting life on other planets.
Researchers have already created alternative amino acids in the lab and added individual alternative amino acids to proteins otherwise made of standard ones. However, no one has built entire proteins from scratch using nothing but a set of alternative amino acids. “This work is genuinely novel,” Freeland says. “Nobody knows what will happen. This is a first.”
Finding the essence
Freeland’s prior work has focused on determining what characteristics are necessary for a set of amino acids to function well as a basis for life. He and his students determined what differentiates our set of 20 amino acids from other random sets of 20 taken from a digital library of about 4,000 mathematically-generated amino acid structures.
That work led to a group of four equations that can be used to describe any set of 20 amino acids. “So it’s almost as if conceptually we boiled it and got the essence of it,” Freeland says, “We identified really unusual, simple characteristics that distinguish life on Earth’s set of 20 amino acids from random.”
With those factors in hand—what Freeland calls “design principles”—his team is now using a powerful computer program they developed to look at other sets of 20 amino acids and find examples that meet the same criteria. And this time around, they are taking the amino acids from a catalog of about 300 that can be obtained commercially—so any “good” set they find can be tested in the lab.
Going back in time to leap forward
This summer, the work entered a new phase. Sean Brown, a UMBC Ph.D. candidate in biological sciences working with Freeland, traveled to the Czech Republic to work in Klara Hlouchova’s lab, where they are preparing the technology to produce proteins with proposed alternative amino acid alphabets. “Basically, we’re asking, ‘Do our design principles work’?” Freeland says.
In some ways, this work will be like going back in time to the mid-20th century, when researchers were first discovering how life behaves at the molecular level. Since then, techniques for building and analyzing proteins have come to rely on everything we’ve learned about life’s amino acid alphabet. As such, these tools cannot be applied to alternative amino acid alphabets. For this project, the team will return to older approaches and techniques, in which the Czech researchers have retained expertise, to see if their alternative amino acids can actually form proteins.
If the experiments are successful, the team will be able to further hone the design principles for a generic life-producing amino acid alphabet, rather than basing the principles solely on the characteristics of life on Earth.
“If we can show that unusual, abstract properties of life as we know it are necessary, it helps us to understand why evolution on Earth went the path it did,” Freeland says. “It’s part of our own history, and it answers the question, ‘Why those 20?’”




Knowing what to look for around the universe
Beyond understanding our own history, refining the design principles would better inform our approach to seeking life on other planets, Freeland says. Scientists generally agree that other life forms are most likely to be microbial, but “most of the ways we currently detect microbes absolutely rely on the molecules in use for life on Earth,” Freeland adds. This work could help researchers know how to detect a “molecular signature” of amino acids that are most likely to show up in life forms that originated independent of Earth.
Like all technologies, these developments come with great promise alongside risk. Freeland isn’t particularly concerned about individual synthetic proteins getting into the wild in the current stage of research, because proteins can’t replicate on their own. But he notes that there are important questions to ask about future, more advanced research. For example, there is potential for scientists working with DNA to build programmable genetic codes. Once this technology merges with fully synthetic proteins, he says, “is where the story takes a really interesting turn.”

Open-source proteins
Possible beneficial applications of synthetic proteins abound, and could include medical treatments or removing toxins from the environment. But without proper oversight, the technology could potentially also lead to harm, Freeland cautions.
Freeland is hoping to avoid the synthetic protein field following in the footsteps of synthetic DNA research. That field is much further ahead, but has mostly been carried out by private companies out of public view, “lending itself to patents and commercial ownership rather than to healthy, transparent debate and legislation,” Freeland says.
To create the best chance for responsible regulation around the use of synthetic proteins, Freeland and his colleagues are trying to quickly get their research into the public domain, he says, to make as robust as possible “what I think will be one of the more important debates of the 21st century when it comes to biotechnology: What are the laws, what are the ethics guiding our use of synthetic proteins and DNA?”
“Life on Earth is an example”
As Freeland and his colleagues lead the way into a new era of synthetic protein research, questions abound. Will proteins form from the amino acid combinations recommended by the algorithm? How can and should we use synthetic proteins, and who will decide? Will the research help us discover life on other planets? As the work develops and more researchers join this burgeoning field, researchers and the public will be constantly reminded of what Freeland describes as “one of the deepest principles of astrobiology.”
“Life on Earth is an example,” he says. “It would seem surprising to think it’s the only example.”

