Scientists understand many of the body’s processes, like breaking down sugars and generating energy for the cell, pretty well. They know what chemical reactions are involved, what molecules they produce, and in what order everything happens. Complex maps even exist of how the different processes interact with one another. There’s a problem, though: the maps are two-dimensional, and cells are three-dimensional. Add the element of time, and you’re up to four dimensions.
Minjoung Kyoung, assistant professor of chemistry and biochemistry, has never been satisfied with 2D maps of 4D processes. “I’ve always been interested in how proteins are working in the real system, in real time, in real action,” she says.
To address the limits of current understanding, Kyoung and her graduate student, Erin Kennedy, ordered parts to build an innovative type of microscope, found in just a few labs around the world. This new tool gave them the rare ability to look at entire living cells at exquisite resolution, as they change in real time. Finally, they could move forward with constructing a 4D map of cellular metabolic pathways.
Kyoung’s preliminary results with the new instrument are promising. Now, with a five-year, $1.6 million grant from the National Institutes of Health, she’s poised to make serious breakthroughs in how we understand the functional relationship between metabolic pathways. Her first targets will include essential basic processes like glucose metabolism (sugar breakdown) and cellular respiration (energy production for the cell, which relies on glucose). Both are fundamental to diseases like diabetes, cancer, and obesity.
Anticipating disease
One thing Kyoung’s early results suggested is that the enzymes important for breaking down glucose and for generating energy are physically close together in the cell—but only when both pathways are functioning normally. “So when they are functionally linked, they are spatially related,” Kyoung says. Her continuing research will try to determine how and why that happens, by looking very carefully at what’s going on in whole cells at various time points and under different cellular conditions.
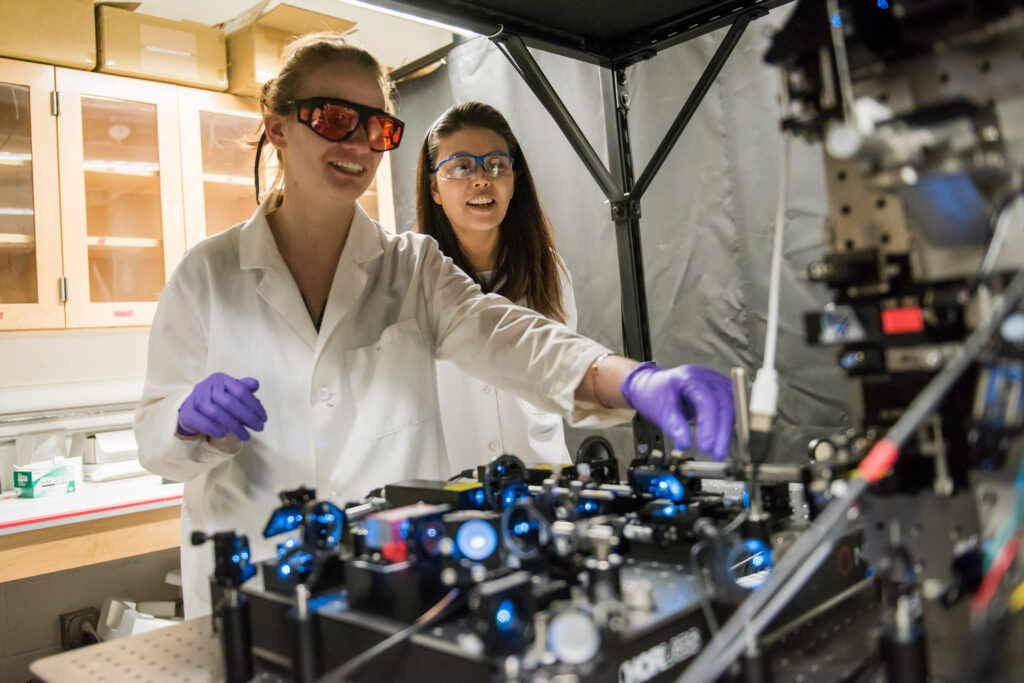
Kyoung also finds the glucose pathway itself to be fascinating. It takes place in the cytoplasm, the watery fluid that fills cells. But somehow, the enzymes required to break down glucose form dense clusters, which Kyoung has dubbed “condensates,” even though the clusters don’t have a formal boundary. “The fundamental mechanism for how these condensates are reversibly assembled and disassembled is one of the specific aims that we’re going to study,” Kyoung says.
The enzymes for the cellular energy pathway also cluster, but they are enclosed inside mitochondria, a structure surrounded by a membrane. A single cell can contain from zero to thousands of mitochondria, depending on the cell’s job. Kyoung explains, “Mitochondria are very important for various metabolic diseases—cancer, diabetes, obesity, and so on. How these mitochondria relate to glucose metabolism is the most important part. So, by understanding them, I truly believe that we can get much, much closer to understanding how these diseases are caused, thus promoting therapeutic intervention.”
“My dream is to be able to predict disease before symptoms occur,” she shares. “That would be the best.”
Ready for a challenge
Getting to the point of recognizing disease before symptoms are apparent won’t be easy. The imaging techniques Kyoung, Kennedy, new graduate student Tao Zhang, and UMBC collaborator Songon An, associate professor of chemistry and biochemistry, are employing are so new, and so difficult, Kyoung anticipates many challenges.
“There is no previous data whatsoever. There is no technical approach whatsoever. There is no approach to data analysis whatsoever,” says Kyoung. She describes being at this cutting edge as both exciting and intimidating. To even successfully collect useful data, “many things have to go right,” Kyoung says.

To see what they want to see inside the cells, such as a particular enzyme, Kyoung’s team will need to tag it with a fluorescent protein, a process that is successful in 50 to 60 percent of cells. That’s not a problem when you use a conventional microscope, because you can see lots of cells at once. But the microscope that enables observing living cells with the resolution Kyoung needs can only see a few cells at a time. So finding the tagged cells has been the first challenge.
After the images are collected, a complex mathematical process called “deconvolution” removes the distortion that the microscope’s light beam itself generates in the images. That takes several hours for a single cell. And then they can actually analyze the images to see which enzymes are where, when. This process takes several days for one cell. Only at that point do they know if the experiment worked.
And, “Because no one has done this type of research before, we have to figure out how we are going to validate our results, too,” Kyoung says. “There is no precedent.” Despite all these challenges, Kyoung is excited to get to work. She believes the kinds of relationships they’ve started to see between glucose metabolism and mitochondria are only the tip of the iceberg as far as spatial relationships between metabolic pathways in the cell.
“Just a start”
“This is just a start. So far we have focused on these two metabolic pathways, but I believe this phenomenon is not limited to just these two,” Kyoung says. “So I envision that this will be the beginning for a big 4D map of all the metabolic networks.”
Kyoung and her team have significant funding from NIH to support their work, the microscope they need to do it, a healthy sense of optimism, and a commitment to helping answer some of the fundamental questions surrounding emerging epidemics like cancers, diabetes and obesity. With the key elements in place, they are bound to make breakthroughs that move the needle on tackling some of today’s most challenging diseases.
Banner image: Minjoung Kyoung and her UMBC lab group. From left to right: Keynon Bell, Minjoung Kyoung, Erin Kennedy, Manuel Huerta-Alvarado, and Tao Zhang. Photo by Marlayna Demond ’11 for UMBC.
Tags: ChemBiochem, CNMS, Research